LC/MS
LC/MS Analysis of Olygonucleotides
Using a Polymer-Based Diol Column - Shodex™ HILICpak™ VN-50 2D Introduction The oligonucleotide therapeutics have high expectations for the usage in not only treating genetic and metabolic disorders, but as for anti-cancer drugs and vaccines of various diseases, because of their unique mechanisms of action. They have abilities to target specifically bind with deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). They also have roles controlling the expression of specific genes that are responsible for certain diseases. The development and quality control of the oligonucleotide therapeutics require highly selective and sensitive analytical methods. The LC/MS measurement is one of them. The most often used method for the analysis of oligonucleotides has been the reversed phase chromatography with the use of ion- pair reagent. However, the ion-pair reagents tend to remain on the LC system and cause several problems. Ion- exchange chromatography is another method commonly used, but it requires using highly concentrated salt in its eluent, which does not work well with the MS detector. The Shodex™ HILICpak™ VN-50 2D column used in this application is a high performance HILIC column, packed with multi-porous polyvinyl alcohol polymers modified with diol functional groups. The 2.0 mm internal diameter of the column is designed to work well with LC/MS measurements. We have been successfully measuring various oligosaccharides using the VN-50 series columns. Now, it is found to be also effective for the analysis of oligonucleotides. This application introduces the methods for analyzing oligonucleotides using the VN-50 2D column with a gradient elution of a volatile basic solvent and acetonitrile. 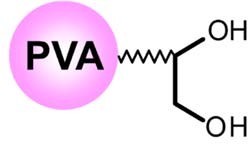 Fig.1 A schematic diagram of VN-50 packing material Experimental There are two types of oligonucleotide therapeutics, categorized in DNA and RNA types. In this application, we used three synthesized DNA oligos (unpurified, salt-free grade) dissolved in ultra-pure water as the test samples. An DNA oligo has a structure having a side chain composed from a combination of four nucleobases; adenine (A), thymine (T), guanine (G), and cytosine (C). Each test sample used were commercially synthesized, supposed to have the base sequences shown below. However, it is likely that all samples contain some fragmented components from the extension interruptions, deletion of bases, and such. Test Sample 1 (20mer): 5’-ATACCGATTAAGCGAAGTTT-3’ Test Sample 2 (20mer): 5’-ATACCAATTAAACAAAATTT-3’ Test Sample 3 (62mer): 5’-ATGAGAAGTATGACAACAGCCCCACACCGGCTGTTGTCATACTTCTCATGGTTCTTCGGAA-3’ The LC/MS system used was Shimadzu Nexera / LCMS-8030 Plus. The column used was Shodexä HILICpak™ VN-50 2D (2.0 mm I.D. x 150 mm; particle size 5 μm; pore size 100 Å). High pressure isocratic or linear gradient with Eluent A - ammonium formate aqueous solution and Eluent B - acetonitrile were used. The flow rate was set at 0.2 mL/min and the column temperature was set at 40 °C. The MS was coupled with a PAD (190-350 nm). An ESI was used as a means of ionization and Scan (-) or SIM (-) mode was used for the detection. The injection volume used was 1 μL.
Results and Discussion
We tested several solvents to choose a suitable Eluent A, including ammonium formate, ultra-pure water, aqueous ammonium hydrogen carbonate, and ammonium water. It showed that the DNA oligo was only retained by ammonium formate, and thus we decided to use it as the Eluent A. Next, we optimized the eluent conditions. Fig.2 shows the UV chromatograms obtained for analyzing the Test Sample 1 using different concentrations of the Eluent A and different ratios of Eluent A/B for the gradient elution. When an isocratic elution of 100 mM ammonium formate with 57% acetonitrile was used, the main component of the Test Sample 1, 20 mer DNA oligo, was detected around 15 minutes. From the MS chromatograms, the DNA oligos smaller than 19 mer were detected before the 20 mer DNA oligo’s peak, eluted in the order of smaller to the larger degree of polymerization. As the degree of polymerization increases, the hydrophilicity increases. Thus, the main separation mode worked in this analysis was assumed to be HILIC. The separation of smaller DNA oligos were not sufficient in this method. To improve their separations, we modified the gradient conditions as such. The initial concentration of the Eluent B (62%) was linearly decreased to 56% over a 10 minute, and the concentration was held for the next 10 minutes. Using this condition provided the most well balanced resolutions and the analysis time. It is preferred to use an eluent with a lower salt concentration to increase the MS’s detection sensitivity. Thus, we tested 20 and 50 mM ammonium formate for the Eluent A. Using 50 mM ammonium formate did not cause any significant influence on the separation nor peak shapes, however, disturbances in the peak shapes were observed when 20 mM ammonium formate was used. Therefore, we decided to use the 50 mM ammonium formate for Eluent A. After the elution of the target DNA oligo, it takes 5 minutes to equilibrate the column back to the initial condition. 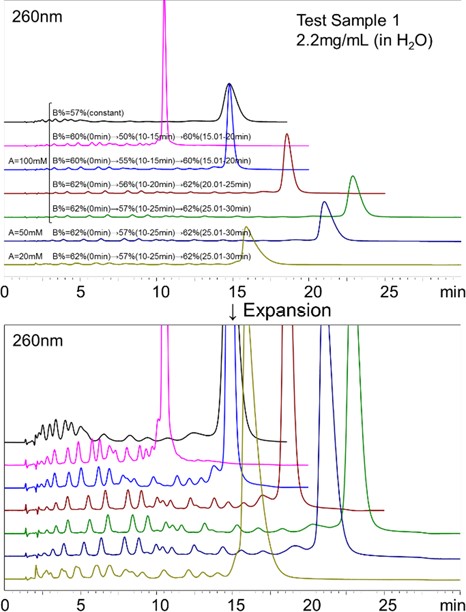 . . Fig.2 Comparison of various eluent conditions for the analysis of the Test Sample 1 1-2. LC/UV/ESI-MS analysis of the Test Samples 1 and 2
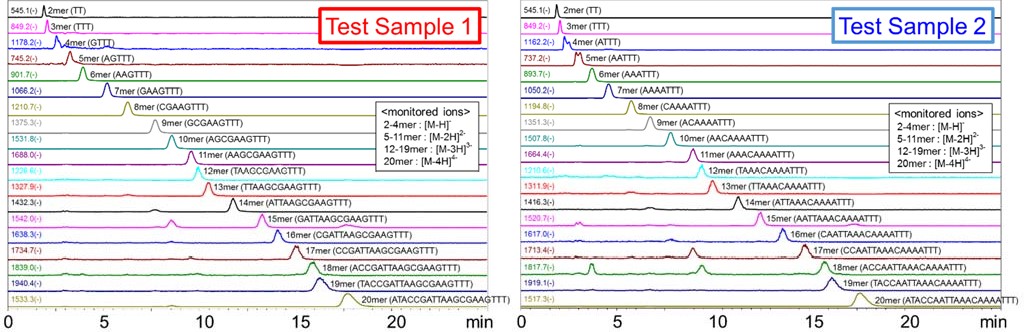
Fig.3 shows the comparison of UV chromatograms (260 nm) for the Test Samples 1 and 2. The gradient condition used was as such. The initial concentration of the Eluent B (62%) was linearly decreased to 56% over a 10 minute, and the concentration was held for the next 10 minutes, and then returned to 62% to equilibrate the column for the next run. The Test Sample 1 which contains the nucleobase G seemed to be retained longer than the Test Sample 2 which does not contain the nucleobase G. It is known that when DNA forms double strands, G and C form stronger base pairs than A and T do. This indicates that hydrogen bonding between G and the packing material has an influence on its retention. 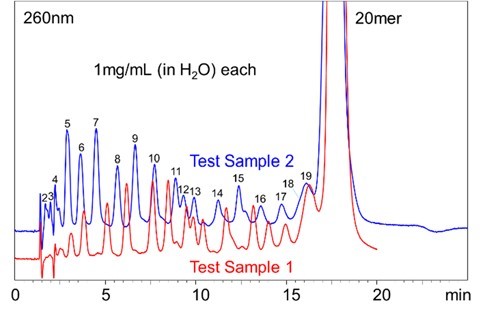 Fig.3 UV Chromatograms of the Test Sample 1 and 2 Fig. 4 shows the Mass spectrum (-) of the 20 mer DNA oligo in the Test Sample 1. It was confirmed that the DNA oligos were detected as multi-valent negative ions formed by losing multiple protons. The multi-valent negative ions on each DNA oligo chains were monitored using the SIM mode. Fig. 5 and 6 show the MS chromatograms of the Test Samples 1 and 2 respectively. In both samples, the DNA oligos of various chain lengths eluted in the order of lower to the higher degree of polymerization. Between some peaks, the resolution of the UV chromatogram was not sufficient to identify each peak (e.g., between 11 and 12 mer or 18 and 19 mer). However, they can be monitored by using the MS chromatograms.
 of the Test Sample 1 (20 mer).jpg) Fig.4 Mass spectrum (-) of the Test Sample 1 (20 mer)
Fig.5 Mass chromatograms of the Test Sample 1 Fig.6 Mass chromatograms of the Test Sample 2 Fig 7 shows the MS chromatogram and the calibration curve of the 20 mer DNA oligo in the Test Sample 1. To shorten the analysis time, the gradient condition was modified as such. The initial concentration of the Eluent B (60%) was linearly decreased to 55% over a 10 minute, and the concentration was held for the next 5 minutes. The linearity of the calibration curve obtained was well. Using the SIM mode, 20 mer DNA oligo can be selectively quantified in the 10 μg/mL level. The analysis time can be shortened even more by decreasing the concentration of acetonitrile. 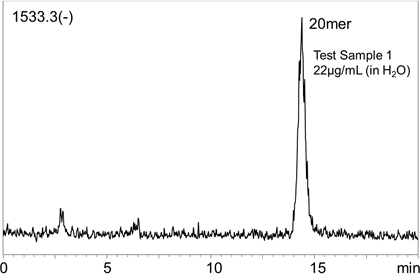 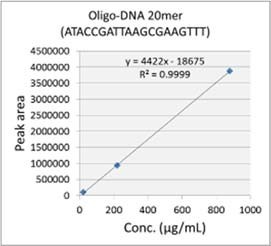 Fig. 7 MS chromatogram and the calibration curve of the Test Sample 1
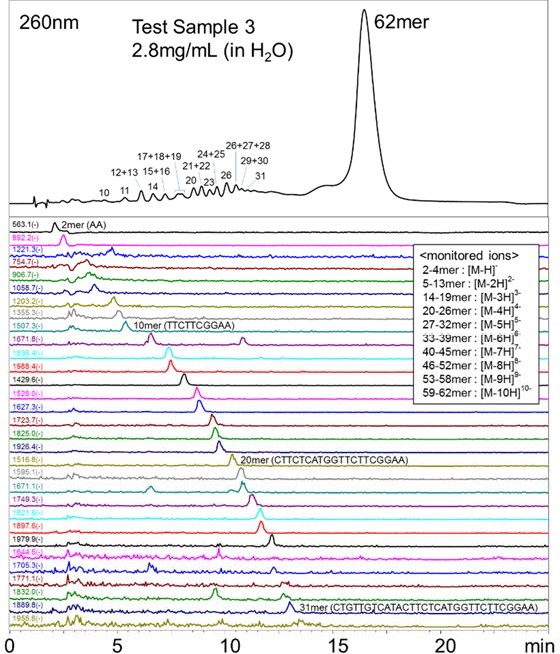
Fig.8 UV and Mass chromatograms of the Test Sample 3 Conclusions This application demonstrated the feasibility of analyzing up to about 30 mer oligonucleotides using the polymer-based diol HILIC column, Shodex™ HILICpak™ VN-50 2D, coupled with an LC/UV/ESI-MS. The method is very selective by separating and eluting the oligonucleotides in the order of lower to higher degree of polymerization. The gradient elution of 50 mM ammonium formate / acetonitrile used in this method was very simple, especially when compared to the commonly used reversed phase chromatography method that requires an addition of ion-pair reagent or to the ion-exchange chromatography method that requires an addition of highly concentrated salts. Moreover, as the ammonium formate / acetonitrile eluent used here is volatile, it has an advantage in simplifying the desalting process during purification. The polymer-based packing material also allows the use of alkaline washing solutions which helps preventing the non-specific adsorption and carry-over related problems that are concerned in the analysis requiring high precision and accuracy or in the preparative scale measurements. Therefore, the methods using the VN-50 series columns introduced in this application have high potentials in the development, quality control, and the purification of oligonucleotide therapeutics. TA.NO.004.①18.D.JUL.P
|
立行科技 Analab Corporation
立行科技 Analab Corporation
| 電話 02-2776-6931 | 傳真 02-2776-6932 | sales@analab.com.tw | |||||
| 台北市中山區長安東路二段181號6樓之2 | |||||||